RESEARCH
Our mission is to advance our understanding of the pathogenesis of human diseases and to streamline the design of personalized therapeutic interventions by using a combination of biological network models (metabolic networks, genetic networks), computational tools (mathematical optimization, machine learning, data mining) and experimentally tractable laboratory systems.
Toward this end, we use these computational and experimental technologies to construct mechanistic models of microbe-microbe, host-microbiota and host-tumor interactions as well as interactions between different human cell lines.
Currently active research areas in the lab include the following:
Host-Microbiota Interactions in Chronic Human Diseases
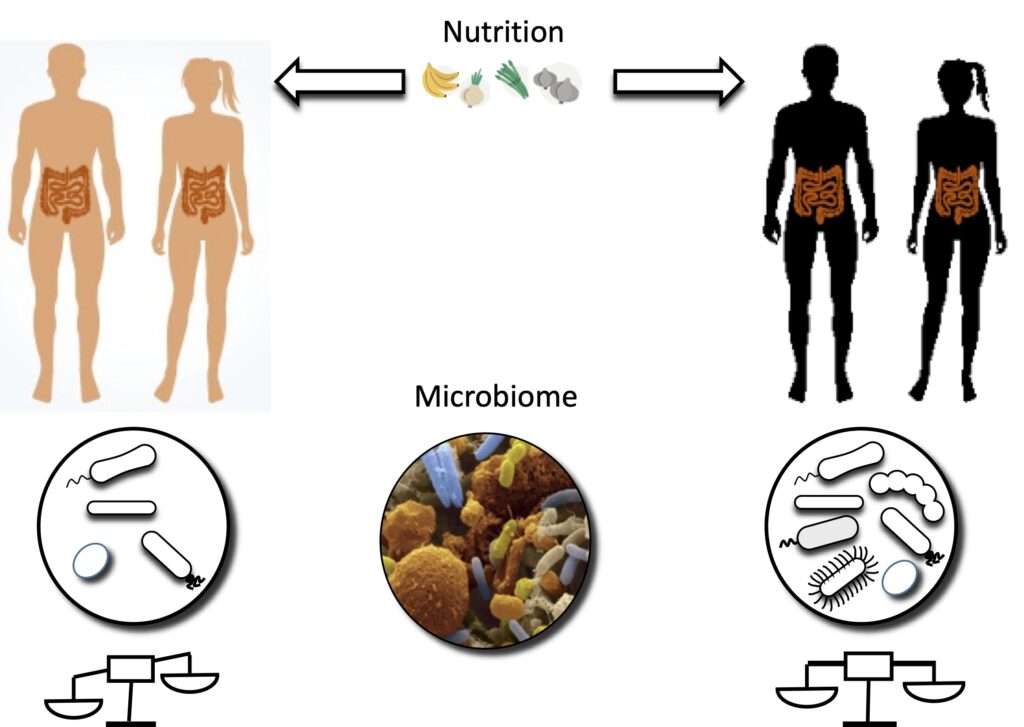
Our lab investigates how intricate interactions between the gut microbiome, the host, and the environment drive the development, progression, or recovery of chronic human diseases. We are particularly interested in the role of metabolism and nutrition in modulating these interactions and shaping immune responses and disease outcomes.and pathogenesis of chronic autoimmune disorders and cancer.
To address these questions, we take an integrated systems approach that combines computational analysis and modeling with experimental systems, including patient-derived 3D gut organoid models. Our research leverages genome-scale models of metabolism for the microbiome and host cell lines/tissues, statistical methods, and advanced mathematical optimization to unravel the complex biological networks underlying host-microbiome crosstalk..
We employ cutting-edge multi-omics technologies, including long-read sequencing, to capture microbial and functional variation with high precision and resolution. Current projects focus on the role of the intestinal and breast milk microbiota in celiac disease, inflammatory bowel disease, and colorectal cancer, with an emphasis on how dietary and microbial metabolites influence host health and disease.
AI for Multi-omics Analysis
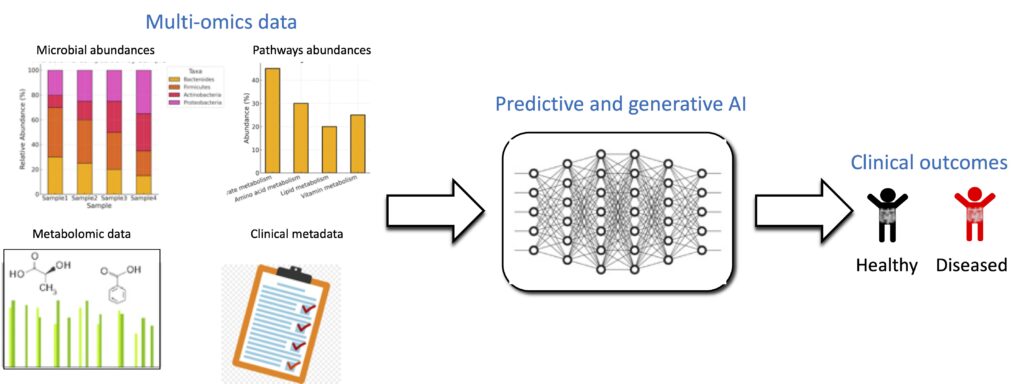
We develop advanced machine learning tools and foundation models to integrate diverse multi-omics datasets—including microbiome, host-derived molecular profiles, and clinical metadata—to enable precise prediction of clinical outcomes. Our work focuses on building predictive and generalizable AI frameworks that can learn from complex, high-dimensional biomedical data, offering scalable solutions for biomarker discovery, disease stratification, and personalized medicine.
Generative AI in Healthcare

We explores how large language models (LLMs) can be harnessed to support clinical decision-making. We evaluate their potential to enhance diagnostic accuracy, summarize complex medical information, and assist healthcare providers in making more informed decisions. In parallel, we rigorously investigate the presence and impact of bias in these models to ensure their safe, equitable, and trustworthy application in healthcare settings. By combining clinical insight with cutting-edge AI, we aim to responsibly advance the integration of LLMs into real-world medical practice.. .
Whole-Body Modeling of Human Metabolism
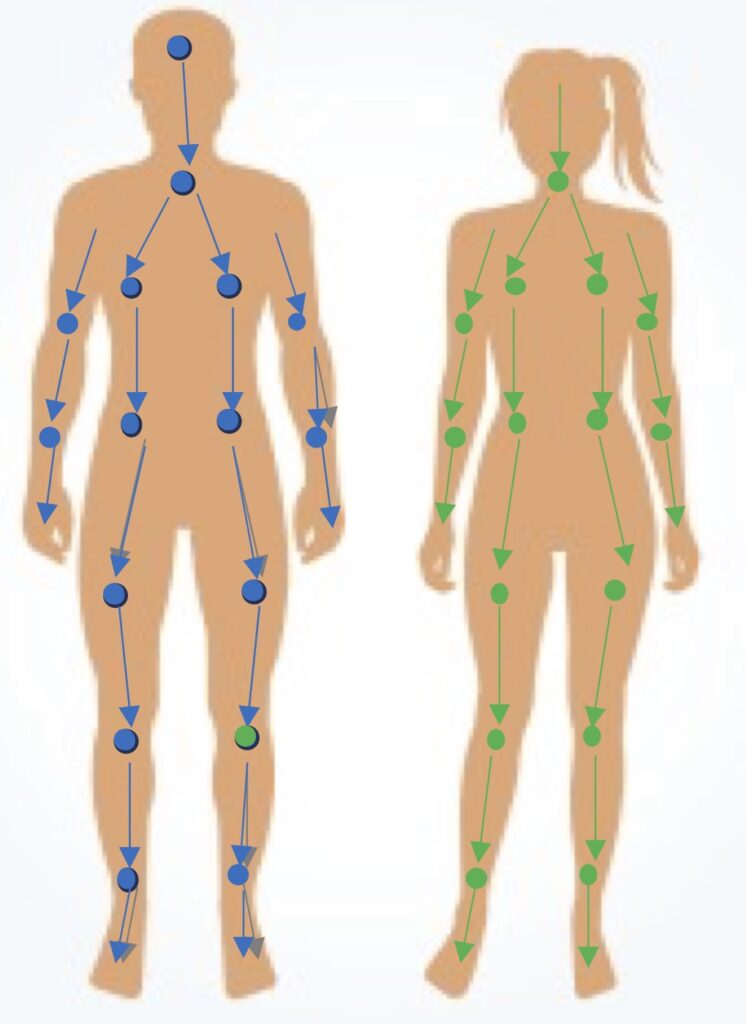
We develop and apply whole-body models of human metabolism to gain a systems-level understanding of how nutrients, metabolic pathways, and organ-organ interactions shape human physiology and disease. These comprehensive models integrate metabolism across multiple tissues and organ systems, enabling us to simulate physiological responses to dietary interventions, metabolic perturbations, and disease states.
Our current research in this area focuses on uncovering sex-specific, tissue-specific, and systemic metabolic features that contribute to complex disorders such as metabolic syndrome and other chronic metabolic diseases. We use these models to generate mechanistic hypotheses, identify biomarkers, predict therapeutic targets, and design personalized dietary recommendations—with the goal of advancing precision nutrition and personalized treatment strategies.
Systems-level Study of the Metabolic Drivers of Immune Response
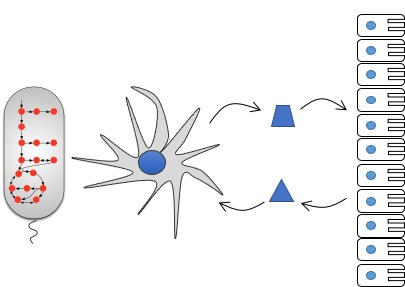
Accumulating evidence suggest that metabolic processes regulate immune cell responses both in healthy subjects and during infection, autoimmune diseases, cancer and obesity. Our lab aims to deepen our understanding of the complex interplay between metabolism and immune responses, ultimately laying the foundation for the development of new therapeutic strategies.
Toward this end, we build genome-scale metabolic network models of key cell types involved in innate and adaptive immunity. These models allow us to simulate immune responses under various physiological and disease conditions, particularly in the context of relevant microenvironments such as the gut. By integrating these models with experimental data, we seek to uncover novel metabolic immunomodulators—metabolites and pathways that can be targeted to either activate or suppress specific immune cell functions..