RESEARCH
Our mission is to advance our understanding of the pathogenesis of human diseases and to streamline the design of personalized therapeutic interventions by using a combination of biological network models (metabolic networks, genetic networks), computational tools (mathematical optimization, machine learning, data mining) and experimentally tractable laboratory systems.
Toward this end, we use these computational and experimental technologies to construct mechanistic models of microbe-microbe, host-microbiota and host-tumor interactions as well as interactions between different human cell lines.
Currently active research areas in the lab include the following:
The role of host-microbiota interactions in chronic autoimmune diseases and cancer
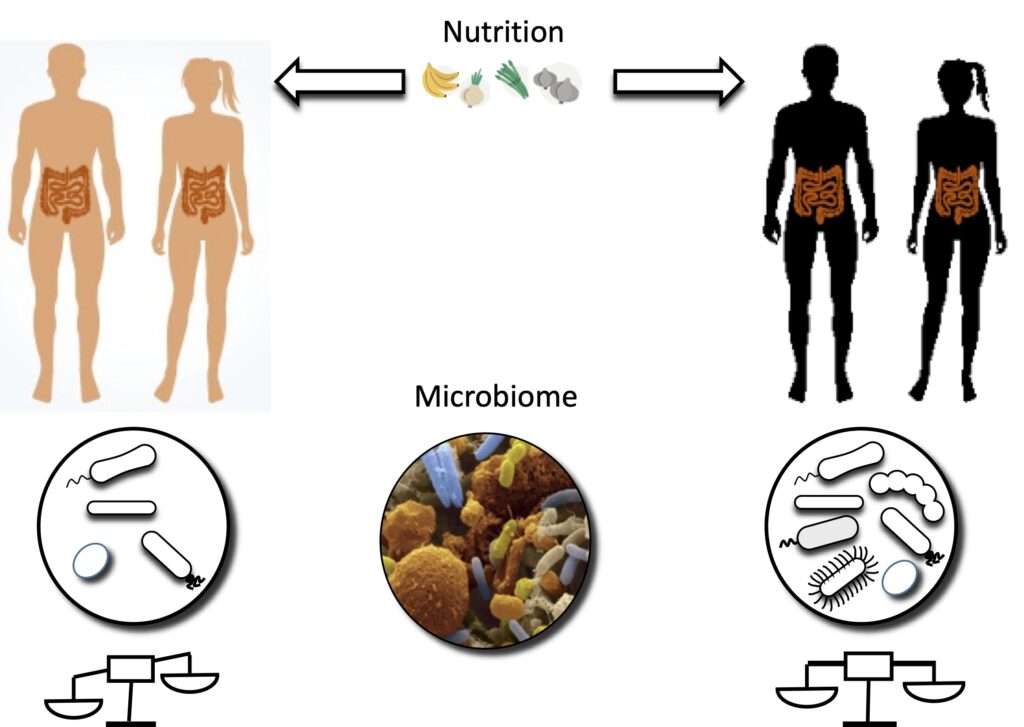
Our goal is to better understand how the way in which microbes interact with each other and with their host and environment contributes to the development and pathogenesis of chronic autoimmune disorders and cancer.
We tackle this question by integrating systems biology approaches, such as genome-scale metabolic network modeling, computational and statistical tools for analyzing large-scale meta’omic (metagenomic, metatranscriptomic, metabolomic) data from microbiome studies and mathematical modeling techniques such as advanced mathematical optimization.
AI in biomedicine
We develop AI-deriven tools that integrate microbiome- and host-derived multi-omrcs data and clinical metadata to precisely predict clinical outcomes.
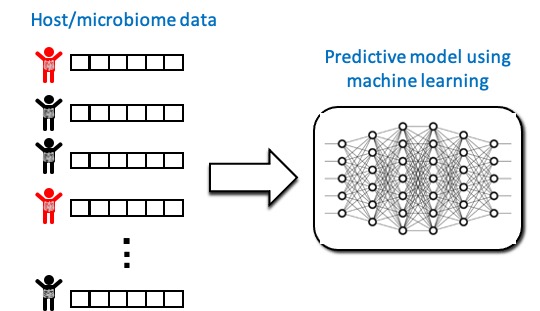
Current projects in the lab focus on the role of intestinal and breast milk microbiota and nutrition in celiac disease, inflammatory bowel disease and colorectal cancer.
Diagnosed with celiac disease? Learn more about the CDGEMM Cohort Study
Long-read sequencing technologies for microbiome profiling
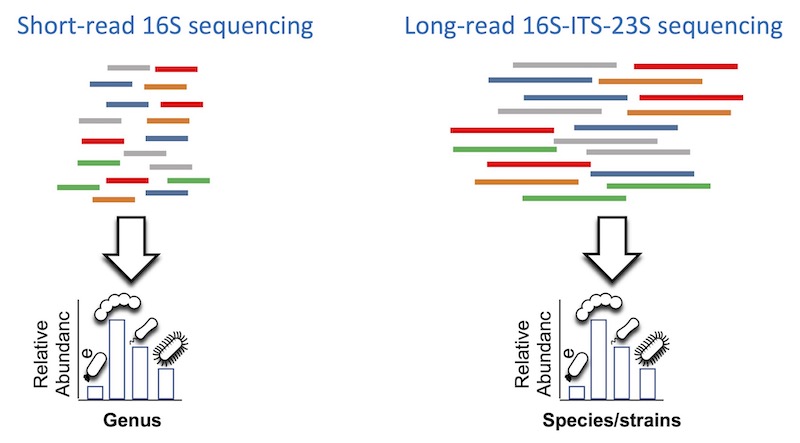
We are interested in utilizing and developing long-read sequencing technologies for the taxonomic and functional profiling of microbiomes. We use both long-read amplicon sequencing and long-read metagenomics to profile microbiomes at species and sub-species resolution and to construct closed genomes for novel strains. Our goal is to shed light on the microbial dark matter in human microbiome studies in ways which is not accessible by using short-term sequencing technologies.
Systems-level study of the metabolic drivers of immune response
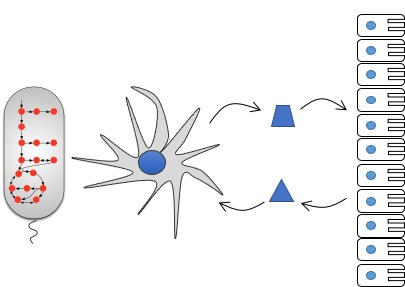
Accumulating evidence suggest that metabolic processes regulate immune cell responses both in healthy subjects and during infection, autoimmune diseases, cancer and obesity. Our goal is to further our understanding of the interplay between metabolism and immune cell response and to provide a foundation for designing new therapeutic targets.
Toward this end, we develop genome-scale metabolic and genetic network models for cells lines involved in innate and adaptive immunity. We use these models to better understand the host immune response to alterations in relevant (e.g., intestinal and tumor) microenvironments and to identify novel metabolic immunomodulator that activate or suppress these immune cells.
The ecology and evolution of cooperation and conflict in microbial communities
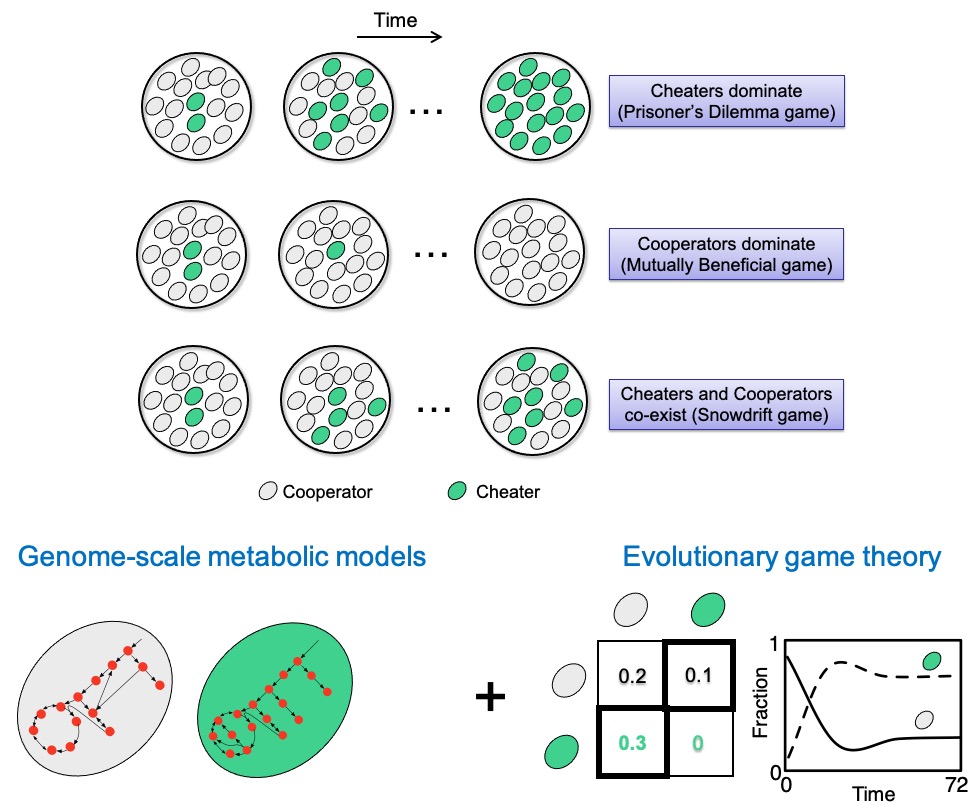
Microbes within microbial communities often interact with each other through the cooperative or selfish utilization of public goods such as dietary carbohydrates and host-derived metabolites. The outcome of these interactions may range from the extinction of a microbial population due to the dominance of a selfish species to evolving toward a stable equilibrium due to the dominance of cooperators or the coexistence of cooperative and selfish species.
We are interested in understanding how these selfish and cooperative interactions will shape the equilibrium and stability of microbial communities, and in finding rational ways of tweaking them toward an equilibrium of biomedical interest.
We address this question by developing hybrid computational and mathematical modeling approaches that integrate genome-scale metabolic network modeling and evolutionary game theory and by designing experimentally tractable laboratory microcosms using model microorganisms such as E. coli or representative microorganisms from the human microbiota.